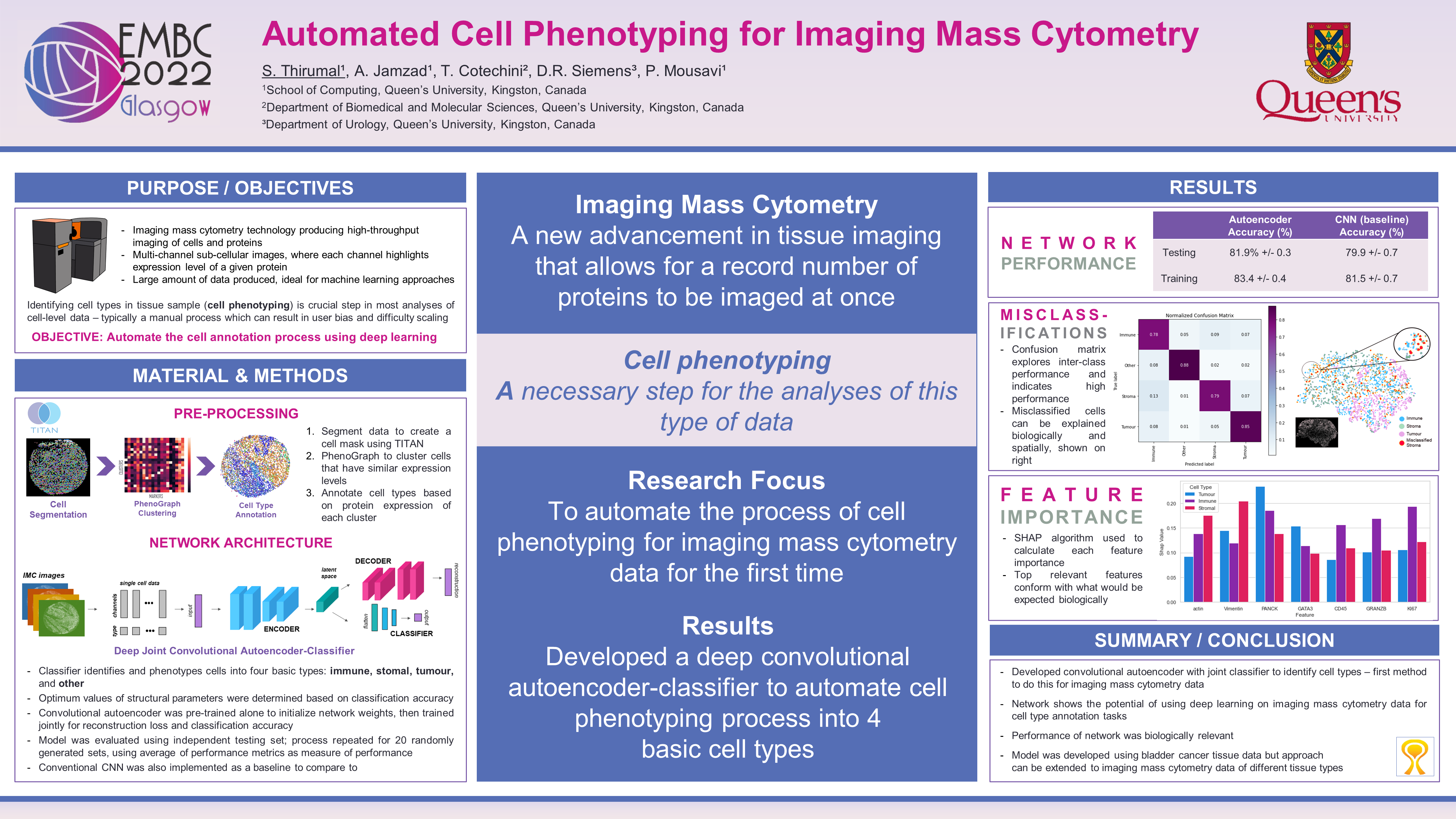
Automating Cell Annotation
Automated cell type annotation using a convolutional autoencoder.
This work was presented and published in 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC).
(click image to open in new window)
Convolutional Autoencoder Structure
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
nFeatures = x_train.shape[1]
model = None
input_ae = Input(shape=(nFeatures, 1))
x = Conv1D(16, 3, activation="relu", padding="same")(input_ae)
x = Conv1D(16, 3, activation="relu", padding="same")(x)
x = MaxPooling1D()(x)
x = Conv1D(32, 3, activation="relu", padding="same")(x)
x = Conv1D(32, 3, activation="relu", padding="same")(x)
x = Dropout(0.7)(x)
encoded = MaxPooling1D()(x)
x = Conv1D(32, 3, activation="relu", padding="same")(encoded)
x = Conv1D(32, 3, activation="relu", padding="same")(x)
x = UpSampling1D()(x)
x = Conv1D(16, 3, activation="relu", padding="same")(x)
x = Conv1D(16, 3, activation="relu", padding="same")(x)
x = UpSampling1D()(x)
decoded = Conv1D(1, 3, activation='sigmoid', padding='same', name='ae_out')(x)
y = Flatten()(encoded)
y = Dense(64, activation = 'relu')(y)
y = Dense(24, activation = 'relu')(y)
output_class = Dense(4, activation = 'softmax', name = "class_out")(y)